Module 3 - DICOM
DAIM Team
Introduction
- Today’s module will cover the DICOM format.
- We will also recap important information from previous seminars
- This will prepare you for the DICOM coding workshop
NumPy Recap
What is a NumPy array?
Why use NumPy arrays for medical images?
- NumPy is built on top of C
- Computations much faster than in Python computations
- NumPy arrays consume less space in memory than Python lists
- NumPy is also fast because it uses broadcasting
Broadcasting
Broadcasting is the ability to perform computations on or using arrays without the need to use for loops even if arrays are not of the same exact shape
- The arrays can be broadcast together if they are compatible with all dimensions.
- If the arrays don’t have the same rank then prepend the shape of the lower rank array with 1s until both shapes have the same length.
- The two arrays are compatible in a dimension if they have the same size in the dimension or if one of the arrays has size 1 in that dimension.
- In any dimension where one array had size 1 and the other array had size greater than 1, the first array behaves as if it were copied along that dimension.

Question
- Considering the rules of broadcasting, which would yield an error and which is compatible?
(2,1), (1,) --->
(5,5,3), (2,5,3) -->
(5,5,3), (5,3) -->
(3,3,4), (4,)-->Image Representation in DICOM
Reminder
- Images are 2D or 3D arrays.
- Image pixels are often represented with 8-bit integers.
- Colour images have three dimensions with a 2D array for every color channel.
- How does this change for medical imaging?

Houndsfield Units
- Used in X-ray imaging (plain films, CT scans)
- They describe a physical property
- The attentuation of x-rays through different structures
- Water is used as a reference point at 0 HU (Hounsfield Units)
- Materials that attenuate X-rays more will have a higher value, and vice versa.

Why is this important for programming?
- Houndsfield units are quantitative - they measure a physical property
- Houndsfield units are typically represented as 12-bit or 16-bit integers.
- As previously mentioned, most images are 8-bit
- How can we visualise CT scans?
Windowing
- Windowing is the selection of a:
- Center HU value (aka level)
- Range (aka window)
- HU values outside of this are not displayed.
- This allows for visualisation of different structures.
| Tissue Type | Hounsfield Unit (HU) Range |
|---|---|
| Air | -1000 HU |
| Fat | -100 to -50 HU |
| Water | 0 HU |
| Soft Tissue | +40 to +80 HU |
| Bone | +700 to +3000 HU |

The DICOM Standard
What is DICOM?
- DICOM (Digital Imaging and Communications in Medicine) is a standard for storing and transmitting medical images.
- It was developed to ensure interoperability between medical imaging devices.
- CT, MRI (stored as 3D images)
- X-ray and ultrasound (stored as 2D images)
- DICOM includes more data about the image itself.
- This is called metadata.
Metadata
- Other formats (JPEG, PNG) have metadata
- DICOM’s metadata is more extensive
- The metadata can include:
- The patient’s name, referring hospital, patient sex etc.
- Paramaters of image aquisition such as slice thickness, protocol, position of patient etc.
Other features of DICOM
- A key part of DICOM is standardization allowing a common framework for managing, storing, and sharing medical images across different devices and platforms
- This allows for integration into electronic health records and PACS (Picture Archive Communication System)
- Support for lossless compression
- Medical images can be compressed without losing valuable scan information
Examples



Break!
Value representation
- Each item of data in a DICOM file has a value representation
- This is its datatype
- Common datatypes in Python include integers, strings, floats, etc.
- DICOM’s value representations are more elaborate
- These are important so that the programmer knows how to interpret data in the file.
Examples of value representations
| VR | Description | Data Type |
|---|---|---|
| PN | Person Name: Stores a person’s name | String (multi-part) |
| DA | Date: Represents a date (e.g., study date) | Date (YYYYMMDD) |
| TM | Time: Represents time (e.g., study time) | Time (HHMMSS) |
| LO | Long String: Limited-length string | String (up to 64 chars) |
| DS | Decimal String: Decimal numbers | String (decimal format) |
| FD | Floating Point Double: 8-byte floating number | 64-bit float |
| UL | Unsigned Long: 32-bit unsigned integer | 32-bit int |
| SQ | Sequence of Items: Represents a sequence of DICOM elements | Sequence (nested data) |
| SH | Short String: Limited length string (up to 16 chars) | String (e.g., hospital name) |
Working with DICOM
DICOM Viewers
- DICOM viewers are specialized software applications that allow users to view and analyse DICOM files.
- Examples of these include:
- OsiriX (mac-OS): Offers advanced visualization and includes metadata extraction features.
- RadiAnt DICOM Viewer: A user-friendly tool that offers detailed views of both image data and associated metadata.
- Horos: An open-source macOS-based viewer with powerful metadata viewing capabilities.
Picture Archiving and Communications Systems (PACS)
- Picture Archive and Communication Systems (PACS) play a crucial role in modern medical imaging.
- They integrate with various imaging modalities and DICOM standards to streamline the management, retrieval, and sharing of medical images.
Key Components of PACS
- Image Acquisition: Receives and stores medical images from different imaging modalities (e.g., MRI, CT, X-ray).
- Data Storage: Maintains a secure, scalable archive for large volumes of image data.
- Image Viewing: Provides user-friendly interfaces for clinicians to access and analyze images.
- Image Distribution: Facilitates the transfer of images and associated data across multiple locations, enabling remote diagnostics and consultations.
What is the difference between PACS and DICOM viewers?
| Feature/Aspect | DICOM Viewers | PACS (Picture Archive and Communication Systems) |
|---|---|---|
| Primary Function | Allows viewing and basic manipulation of DICOM images. | Comprehensive system for storing, retrieving, and distributing medical images. |
| Data Storage | Typically does not provide significant storage capabilities. | Provides extensive, scalable data storage solutions. |
| Image Management | Limited to individual or small sets of images. | Manages large volumes of images across multiple modalities. |
| Integration | Basic integration with imaging devices. | Integrates with various imaging modalities and hospital information systems. |
| Collaboration | Limited to local use, minimal sharing capabilities. | Facilitates remote access, sharing, and collaborative analysis. |
| Accessibility | Often standalone or desktop-based applications. | Web-based or server-based with multi-user access. |
| Cost | Generally more affordable or free. | Higher cost due to advanced features and infrastructure. |
| Advanced Tools | Basic tools for image adjustments and measurements. | Advanced diagnostic tools and analytics for image review. |
| Use Case | Suitable for individual practitioners or small practices needing basic image review. | Ideal for hospitals, large clinics, and healthcare networks that require comprehensive image management. |
Python Packages
- Python can be used to interact with DICOM files
- The library that we will be using in this course is pydicom.
- This package can be used to open and extract image data.
- This package also allows users to view metadata.
- Other libraries include:
- dicompyler: Useful for radiation therapy data analysis and reviewing treatment plans.
Voxels in DICOM
What are Voxels?
- Voxel (short for “volumetric pixel”) is the 3D counterpart of a pixel.
- In medical imaging, voxels represent a volume of tissue in a scanned 3D space.
- Each voxel contains a value that corresponds to the measured property of that volume, such as tissue density or intensity.
Voxel dimensions
- Voxel size depends on the spatial resolution of the imaging technique and the slice thickness.
- Smaller voxels provide higher resolution images but may require more data and longer scan times.
- The size of a voxel is determined by:
- Slice thickness (z-axis)
- Pixel size in the x-y plane
Isotropic vs. Non-Isotropic Voxels
- Isotropic voxels have equal dimensions, e.g. 1 mm × 1 mm × 1 mm
- Non-isotropic voxels do not have square dimensions, e.g. 1mm × 1mm × 2mm
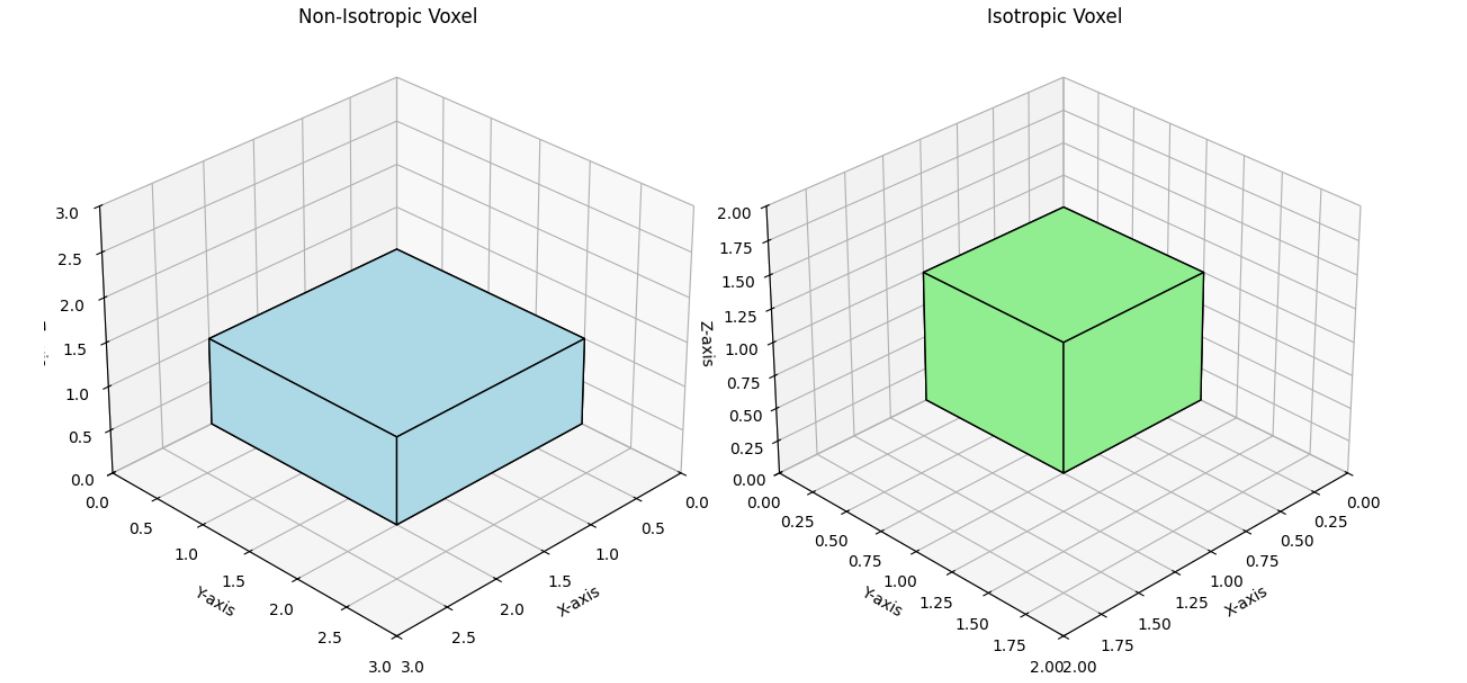
Example plot showing different voxel dimensions.
DICOM Tags for Voxel Dimensions
- Pixel Spacing (0028,0030)
- This tag defines the physical distance in millimeters between the center of each pixel in the image plane (usually in the X and Y dimensions).
- Slice Thickness (0018,0050):
- This tag indicates the thickness of each slice in millimeters along the Z-axis.
Break!
Rendering DICOM Data
Rendering Data
- A 3D image cannot be shown on a 2D screen.
- How do you display data from a 3D volumetric scan?

What the voxels in a chest CT scan look like when plotted as a volume - this is not useful.
Displaying an image
- Sections of an image can be displayed.
- These can correspond to the x, y, and z imaging dimensions
- Coronal, sagittal and axial planes
- In Python, these can be extracted by array slicing
- What happens if you want a different angle of your imaging plane?

Scrolling down the z-axis (axially) in a thoracic CT scan
Interpolation
- Interpolation is a method used to estimate an unknown value between two known values.
- In medical imaging, it can be used to estimate intermediate values between voxels
- This can be useful if you want to section the volume at an angle.
Segmentation
- Segmentation is pixel-level classification where each pixel is classified to belong to a certain class
- e.g. is this pixel part of the liver?
- This can be useful in many settings including automated image analysis and object rendering

Segmentation of the liver and other vessels.
Maximum Intensity Projection (MIP)
- Maximum Intensity Projection (MIP) is a method used to visualize 3D volumetric data
- It projects the highest intensity values along each line of sight onto a 2D plane.
- Often used in positron emission tomography (PET) imaging.

MIP of a SPECT image of a mouse. Source: Christian Lackas, delta@lackas.net Originally uploaded to en.wikipedia. Description page is/was here. CC BY-SA 3.0
Thank you!
Any questions?